TeachOpenCADD
Open source data and software are increasingly generated, developed and used in computer-aided drug design (CADD). This development allows to build modular pipelines for reproducible and reusable research as well as to explore and contribute to open software code. While code and usage of such software is usually well documented, its full potential for CADD projects often remains unreached, especially for beginners, due to the lack of application examples combining different toolkits.
TeachOpenCADD is a teaching platform offering tutorials on central topics in cheminformatics and structural bioinformatics. The tutorials contain theoretical background and practical implementations using open source data and software. Implementations are available in two formats: On the one hand, interactive Jupyter notebooks demonstrate how to set up code-based pipelines (Python). On the other hand, the same topics are transformed into KNIME workflows, an alternative to code-based workflows. Here, an intuitive, drag-and-drop style graphical interface is used to string together pre-implemented code units (nodes) with standardized functionalities.
TeachOpenCADD is suitable for self-study training and classroom teaching, but can also serve as a starting point in research projects. The platform is freely available on GitHub and open to contributions from the community.
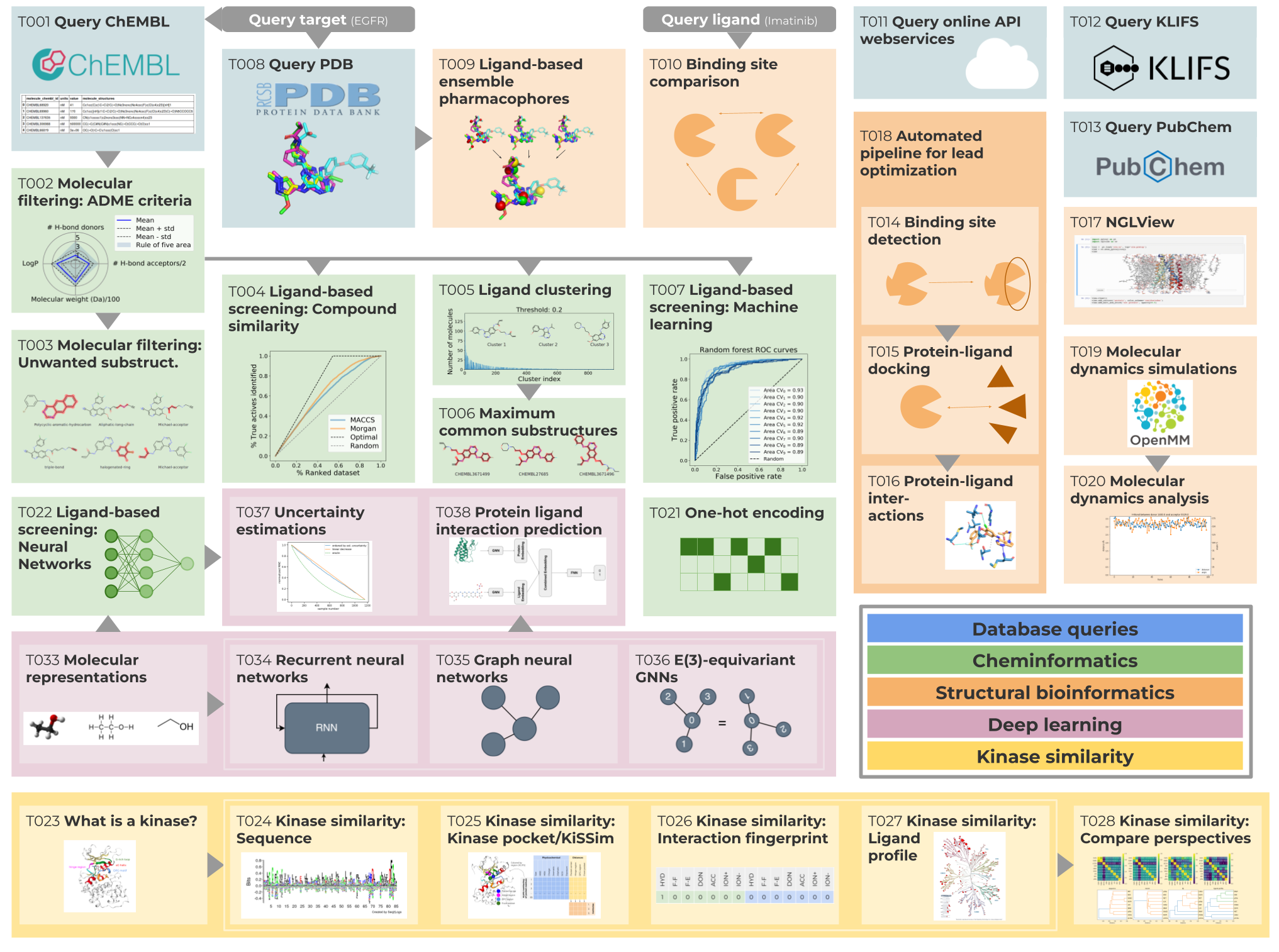
Figure: The TeachOpenCADD platform offers tutorials covering a step-by-step pipeline to propose novel EGFR kinase inhibitors with concepts from cheminformatics (green), structural bioinformatics (orange) and online webserver queries (blue). Talktorials also cover different kinase similarity measures (yellow) and an introduction to deep learning (purple).
Software and resources
- TeachOpenCADD website · Main website for the TeachOpenCADD platform
- TeachOpenCADD for Jupyter · Jupyter notebooks on computer-aided drug design tasks using open resources
- TeachOpenCADD for KNIME · KNIME workflows on computer-aided drug design tasks using open resources
People
- Dominique Sydow
- Jaime Rodríguez-Guerra
- Andrea Volkamer
- Talia B. Kimber
- David Schaller
- Corey Taylor
- Yonghui Chen
- Michele Wichmann
- Mareike Leja
- Sakshi Misra
- Andrea Morger
- Maximilian Driller
- Armin Ariamajd
- Michael Backenköhler
- Paula Linh Kramer
- Floriane Odje
- Hamza Ibrahim
- Greg Landrum · KNIME
- Daria Goldmann · KNIME
- Joschka Groß
- Gerrit Großmann
- Roman Joeres
- Azat Tagirdzhanov
- Verena Wolf
Funding
- Note that the TeachOpenCADD project has been a group effort and has received no explicit funding, while the positions of individual authors were supported by diverse funding agencies, see the individual projects’ pages.
Publications
-
- Michael Backenköhler
- Paula Linh Kramer
- Joschka Groß
- Gerrit Großmann
- Roman Joeres
- Azat Tagirdzhanov
- Dominique Sydow
- Hamza Ibrahim
- Floriane Odje
- Verena Wolf
- Andrea Volkamer
-
- Dominique Sydow
- Jaime Rodríguez-Guerra
- Talia B. Kimber
- David Schaller
- Corey Taylor
- Yonghui Chen
- Mareike Leja
- Sakshi Misra
- Michele Wichmann
- Armin Ariamajd
- Andrea Volkamer
-
- Dominique Sydow
- Michele Wichmann
- Jaime Rodríguez-Guerra
- Daria Goldmann
- Gregory Landrum
- Andrea Volkamer
- (2019). TeachOpenCADD: a teaching platform for computer-aided drug design using open source packages and data. Journal of Cheminformatics 11(29). DOI: 10.1186/s13321-019-0351-x.
- (2021). Teaching Computer-Aided Drug Design Using TeachOpenCADD. Teaching Programming across the Chemistry Curriculum. ACS Symposium Series 1387. DOI: 10.1021/bk-2021-1387.ch010.