Ratar
How to probe and validate a potential target remains one of the key questions in basic research in life sciences. In the DFG-project Ratar (Read-Across the Targetome), we will use binding site similarity to predict off-targets and to extrapolate compound information from one target to another. This similarity-based knowledge transfer can suggest tool compounds (chemical probes) and off-targets for proteins of interest using the ever-growing amount of available target and compound data.
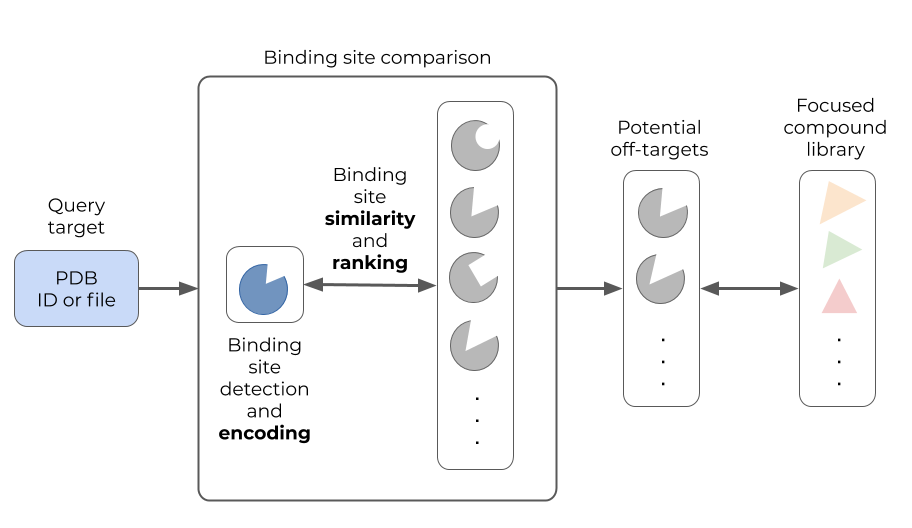
Figure: Read-across the targetome (Ratar) aims to support off-target or polypharmacology studies for a query target using (structural) binding site comparison. Known binders of the most similar binding sites are proposed as tool compounds for the query target.
Software and resources
- ratar · Binding site comparison tool (WIP)
People
Funding
- Bundesministerium für Bildung und Forschung, grant ID 031A262C
- Deutsche Forschungsgemeinschaft (DFG), grant ID VO 2353 / 1-1
Publications
-
- Dominique Sydow
- Lindsey Burggraaff
- Angelika Szengel
- Herman W. T. van Vlijmen
- Adriaan P. IJzerman
- Gerard J. P. van Westen
- Andrea Volkamer