Maxsmi
Deep learning requires lots of data which in the case of physico- chemical and bioactivity remains scarce. Here, we exploit that one compound can be represented by various SMILES strings as means of data augmentation and we explore several augmentation techniques. The best strategies lead to the Maxsmi models, the models that maximize the performance in SMILES augmentation. These models are trained on four data sets, including experimental solubility, lipophilicity, and bioactivity measurements, and are available for prediction on novel compounds.
Moreover, the uncertainty of the models is assessed by applying augmentation on the test set. Our results show that data augmentation improves the accuracy independently of the deep learning model and of the size of the data.
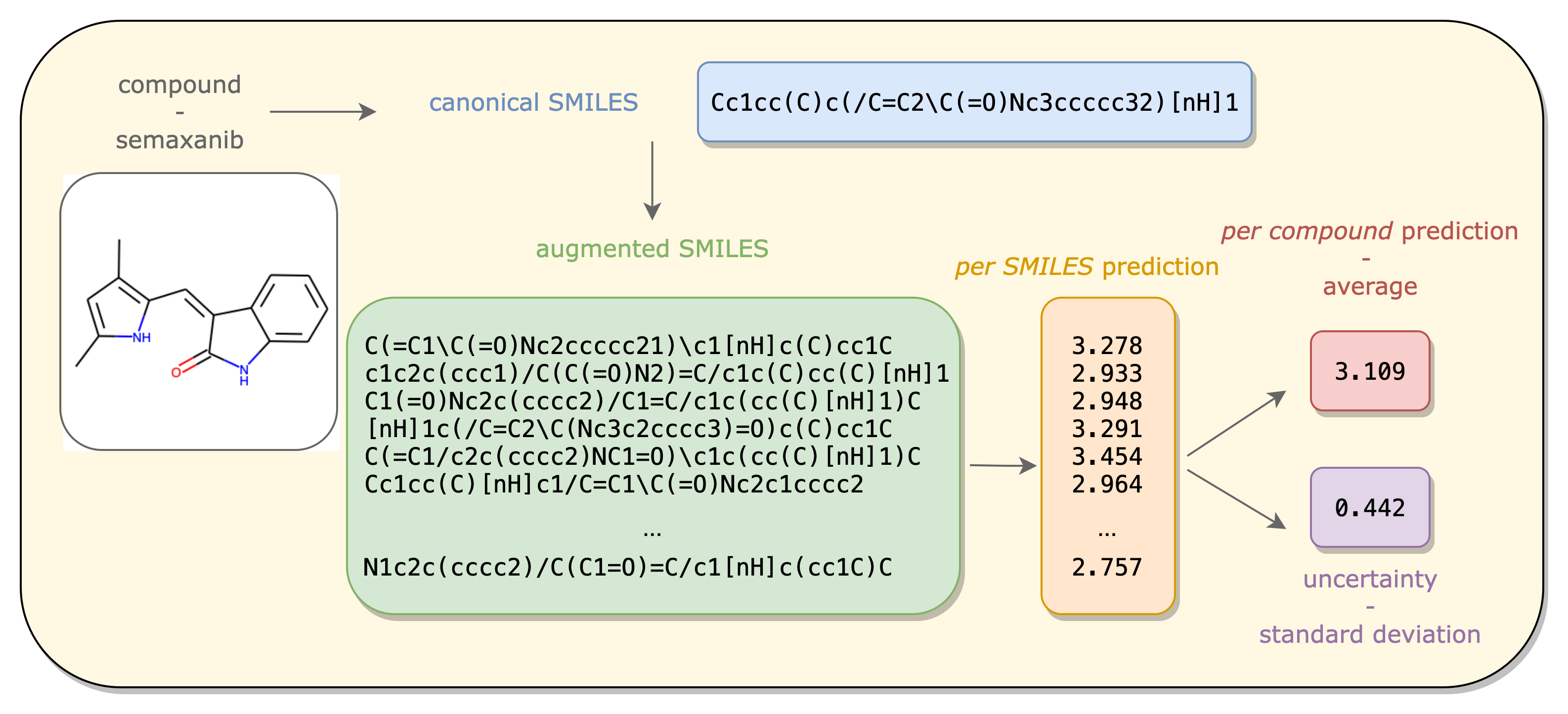
Figure: Given a compound represented by its canonical SMILES, the Maxsmi model produces a prediction for each of the SMILES variations. The aggregation of these values leads to a per compound prediction and the standard deviation to an uncertainty in the prediction. The Maxsmi model predicts lipophilicity of semaxanib to 3.109, with an uncertainty of 0.442. The figure is taken from Kimber, 2021.
Software and resources
- maxsmi · Data augmentation for molecular property prediction using deep learning
People
- Maxime Gagnebin
Funding
- The Einstein Foundation & Stiftung Charité · BIH Einstein Visiting Fellowship
Publications
-
- Talia B. Kimber
- Maxime Gagnebin
- Andrea Volkamer