KiSSim
Kinases are important and well studied drug targets for cancer and inflammatory diseases. Due to the highly conserved structure of kinases, especially at the ATP binding site, the main challenge when developing kinase inhibitors is achieving selectivity, which requires a comprehensive understanding of kinase similarity. In our KiSSim (Kinase Structural Similarity) project, we developed a subpocket-focused kinase fingerprint to investigate kinome-wide pocket similarity.
The KiSSim fingerprint is based on the KLIFS pocket alignment, which defines 85 pocket residues for all kinase structures. This enables a residue-by-residue comparison of spatial and physicochemical features without the need for a computationally expensive alignment step. We aim to use the KiSSim method to detect potential off-targets at an early stage of inhibitor design and to conduct structure-informed polypharmacology studies.
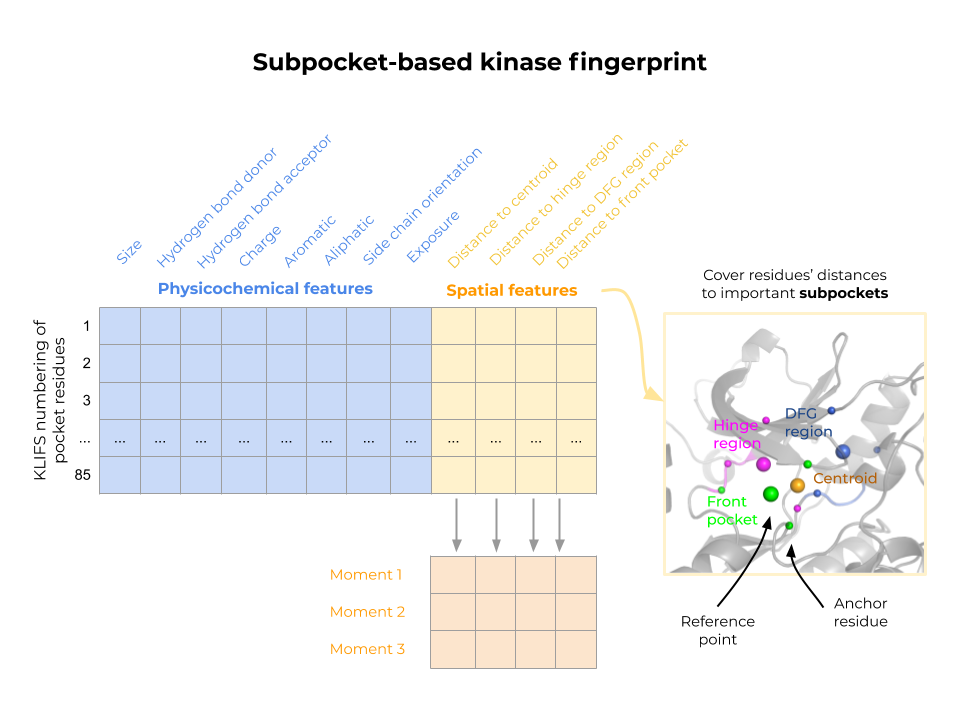
Figure: Kinase structural similarity (KiSSim) is calculated by pairwise comparisons of subpocket-based kinase fingerprints, considering each pocket residue’s physicochemical properties and spatial arrangement towards important subpockets and the pocket center.
Software and resources
-
kissim · Subpocket-based fingerprint for structural kinase comparison -
kissimpackage -
kissim · Subpocket-based fingerprint for structural kinase comparison -
kissimdocumentation - kissim_app · All-against all comparison of structurally covered kinases using the KiSSim fingerprint.
People
- Albert Kooistra · University of Copenhagen
- Friedrich Rippmann · Merck
Funding
- Bundesministerium für Bildung und Forschung, grant ID 031A262C
- Deutsche Forschungsgemeinschaft (DFG), grant ID VO 2353 / 1-1