KinoML
In this project, we aim to combine structure-enabled machine learning and alchemical free energy calculations to develop a predictive quantitative model to rapidly assess kinase inhibitor affinity and selectivity, design ligands with desired selectivity profiles and assess the impact of clinical point mutations on inhibitor binding.
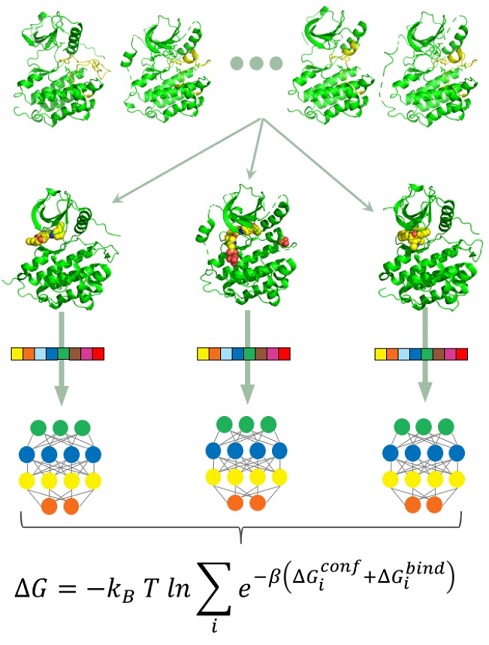
Figure: Small inhibitors targeting kinases can have a wide scope of activity given the conserved ATP-binding pocket across the kinome. Using structure-informed machine learning, we intend to decode the interactions driving the design of promising compounds with intended polypharmacology.
The resulting kinoml package provides a modern Python library to help build flexible pipelines for machine learning in the context of structural bioinformatics. More information on how to use install it and use it for your research can be found on the official documentation.
Software and resources
People
- John D. Chodera · MSKCC
Funding
- The Einstein Foundation & Stiftung Charité · BIH Einstein Visiting Fellowship