KinFragLib
The KinFragLib project allows to explore and extend the chemical space of kinase inhibitors using data-driven fragmentation and recombination, built on available structural kinome data from the KLIFS database for over 2,500 kinase complexes. The computational fragmentation method splits known non-covalent kinase inhibitors into fragments with respect to their 3D proximity to six predefined subpockets relevant for binding.
The resulting fragment library consists of six subpocket fragment pools with over 7,000 fragments and offers two main applications:
- In-depth analyses of the chemical space of known kinase inhibitors allow to investigate subpocket characteristics and connections.
- Subpocket-informed recombination of fragments enumerates new combinations of known fragments in order to generate potential novel inhibitors.
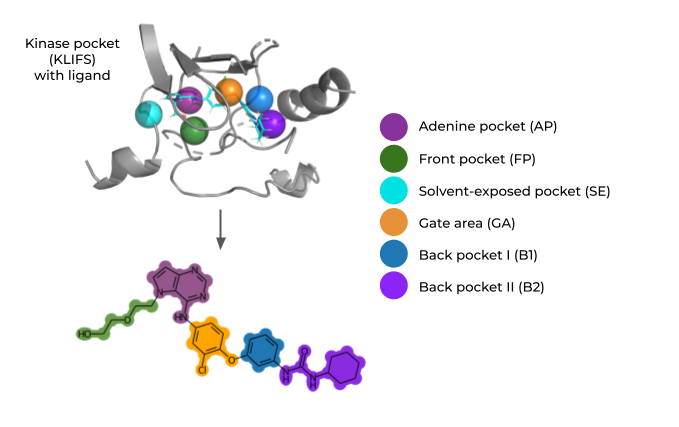
Figure: Ligands co-crystalized with kinases are fragmented with respect to important kinase subpockets, resulting in a kinase-focused fragment library (KinFragLib) used for chemical space analysis and recombination.
Software and resources
- KinFragLib · A kinase-focused fragment library
People
Funding
- Bundesministerium für Bildung und Forschung, grant ID 031A262C
- Deutsche Forschungsgemeinschaft (DFG), grant ID VO 2353 / 1-1
Publications
- (2020). KinFragLib: Exploring the Kinase Inhibitor Space Using Subpocket-Focused Fragmentation and Recombination. Journal of Chemical Information and Modeling. DOI: 10.1021/acs.jcim.0c00839.