Cytotoxicity
In drug development, late stage toxicity issues of a compound are the main cause of failure in clinical trials. In silico methods are therefore of high importance to guide the early design process to reduce time, costs and animal testing.
In this project, cytotoxicity prediction, one of the earliest handles in drug discovery, is investigated using a deep learning approach trained on a highly consistent in-house data set of over 34,000 compounds from the FMP. Albeit yielding good results, neural networks are often described as a black box lacking deeper mechanistic understanding of the underlying model. To overcome this lack of interpretability, a Deep Taylor Decomposition method is investigated to identify substructures that may be responsible for the cytotoxic effects, the so-called toxicophores.
This approach could be helpful in drug development to predict the potential toxicity of a compound as well as to generate new insights into the toxic mechanism. Moreover, it could also help to de-risk and optimize compounds.
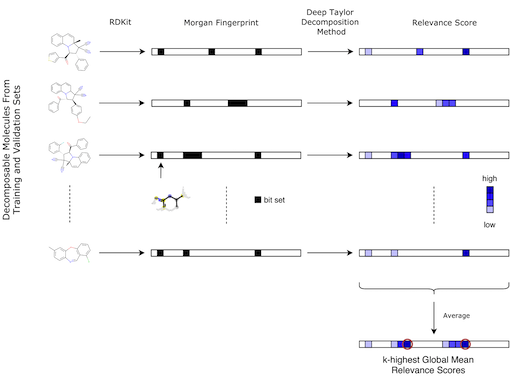
Figure: Workflow for identifying potential toxicophores, taken from Webel, 2020. The first arrow describes the transformation from the molecules in the training and validation sets into 2048 long binary vector describing the Morgan fingerprints of radius 2, using RDKit. Each bit represents one (or more) atom environment(s). The black box indicates if the corresponding atom environment is present in the molecule. The second arrow shows that relevance scores can be obtained for each compound using the Deep Taylor Decomposition method. Once all relevance scores are computed for each decomposable molecule, they are averaged. The bits corresponding to the k-highest global mean relevance scores are stored and used for further analysis as potential toxicophores.
People
- Silke Radetzki · FMP
- Martin Neuenschwander · FMP
- Marc Nazaré · FMP
Funding
- Bundesministerium für Bildung und Forschung, Grant 031A262C
- Stiftung Charité (Einstein BIH Visiting Fellow Project)
Publications
-
- Henry Webel
- Talia B. Kimber
- Silke Radetzki
- Martin Neuenschwander
- Marc Nazaré
- Andrea Volkamer